The Biophysical Society's Annual Art of Science Image Contest took place online this year, during the 65th Annual Meeting. The third place winning image was submitted by Melanie König, a PhD student in the group of Siewert-Jan Marrink at the University of Groningen. König took some time to provide information about the image and the science it represents.
How did you compose this image?
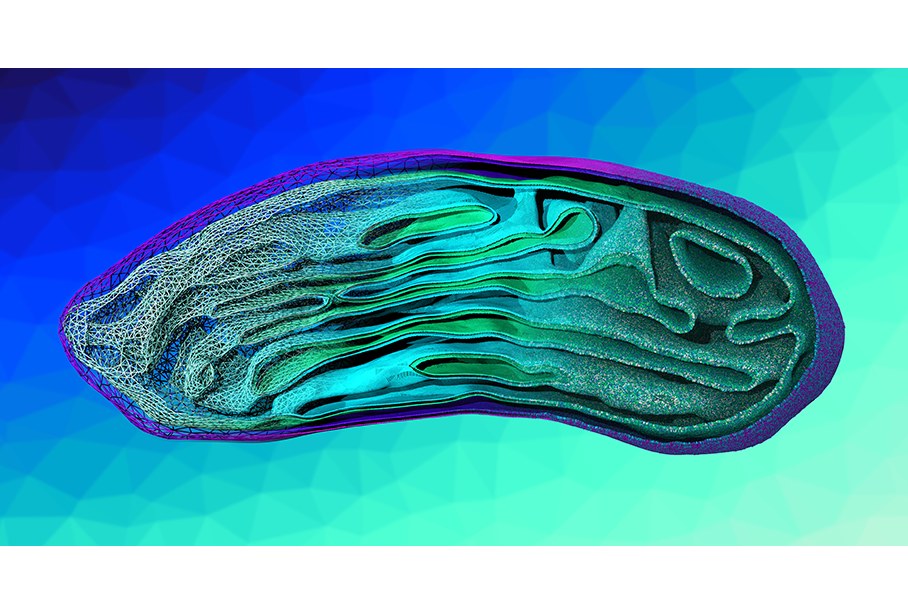
The image of the mitochondrion illustrates our recently developed hierarchical modeling framework (https://doi.org/10.1038/s41467-020-16094-y). Within our multiscale approach, we combine different simulation techniques thereby serving as a computational microscope. The final image is composed of three separate images, all rendered with VMD and smoothly blended together using Affinity Photo. From left to right more information is included in the mitochondrion model: (Left) Based on EM tomography data we built a triangulated surface mesh of the inner (cyan) and outer (violet) mitochondrial membrane. (Middle) In the next step, we generated the corresponding monolayers. Using several pointillism operations separately on the inner and outer membrane, we increased the number of points (inner membrane: green and cyan; outer membrane: magenta and violet). (Right) These points were subsequently used to place a realistic mixture of mitochondrial coarse-grained lipids. The final backmapping level displays a near-atomistic model of an entire organelle with a realistic size (341 x 488 x 791 nm3) after a 2 ns simulation using the Dry Martini forcefield.
What do you love about this image? Or, what about this image made you submit it for the contest?
The image in a way embodies the various scientific disciplines I encounter every day in my field of research. When talking with my colleagues, I notice that each one of them has a different understanding of e.g., a mitochondrion, depending on their scientific background. While a biologist may see a mitochondrion as the powerhouse of a cell, providing the chemical energy that is needed to power numerous vital biochemical reactions, a physicist may focus on the underlying forces e.g., spontaneous curvature and bending elasticity, that determine the shape of the mitochondrion. I as a chemist would describe the mitochondrion as a complex construct defined by molecular interactions between lipids and proteins which determine the lateral membrane heterogeneity. Like the various disciplines, our multiscale approach describes a mitochondrion at different scales. Therefore, it can also serve as a tool to communicate between different scientific fields.
What do you want viewers to see/think when they view this image?
While we chose a mitochondrion to showcase our work, I would like viewers to appreciate that the image is not only about the mitochondrion, but about our backmapping algorithm and multiscale approach in general. In fact, this method could be used for any large biological entity in the same fashion.
How does this image reflect your scientific research?
Our multiscale framework has been developed to study large-scale membrane remodeling processes, like the initial step of vesicle fission and membrane bud formation upon Shiga toxin binding. Recently, we are using the multiscale modeling approach to study the assembly of membrane-anchored FtsZ proteins, which form the so-called Z-ring, leading to constriction of cellular membranes. The formation of a contractile Z-ring is a crucial step in bacterial cell division. To elucidate the underlying mechanisms, we employ molecular dynamics simulations at different spatiotemporal scales. A combination of all-atomistic, coarse-grained and DTS simulations allows us to investigate the system from the interaction of a single FtsZ protein with cellular membranes to large-scale membrane deformations upon FtsZ polymerization.
How might your research be relevant to those who are not working in your specific field?
My research is part of a national research initiative in the Netherlands, called Building a Synthetic Cell (BaSyC). Within that initiative, researchers from diverse fields aim to construct a synthetic cell that is as simple as possible but still comprises all the essential properties of life. Hence, we need to understand the complex interplay between cellular components. This will inevitably provide a deeper understanding of the spatio-temporal dynamics of cellular processes, which may benefit people from various disciplines.
Do you have a website where our readers can view your recent research?
I am a PhD student in the group of Prof. Siewert-Jan Marrink. Check out http://cgmartini.nl for recent developments and applications of the Martini forcefield. I am also part of the Building a Synthetic Cell (BaSyC) consortium in the Netherlands. If you want to know more about how to build life from scratch in a bottom-up approach, visit https://www.basyc.nl.