ROCKVILLE, MD – Coronaviruses exploit our cells so they can make copies of themselves inside us. After they enter our cells, they use our cell machinery to make unique tools of their own that help them generate these copies. By understanding the molecular tools that are shared across coronaviruses, there is potential to develop treatments that can not only work in the current COVID-19 pandemic, but in future coronavirus outbreaks as well. Rockefeller University researchers in the labs of Tarun Kapoor and Shixin Liu, including postdoctoral associate Keith Mickolajczyk, recently published their study of one of these molecular tools, which is a potential drug target. They will present their research on Tuesday, February 23 at the 65th Annual Meeting of the Biophysical Society.
During a viral infection, viruses make copies of themselves inside their host, and viruses carry genetic instructions for several tools in order to do so. One of those tools is called a helicase—all organisms have helicases that unwind the genetic information so it can be read or copied. Mickolajczyk had been studying helicases and other molecular motors when the COVID-19 pandemic hit and turned his attention to a helicase encoded in the genome of SARS-CoV-2 (the virus that causes COVID-19), called nsp13.
Mickolajczyk and colleagues investigated the mechanism that individual nsp13 molecules use to unwind genetic material, and their study marks the first single-molecule unwinding experiments to ever be done on a coronavirus helicase. They found that nsp13 is a relatively weak helicase, meaning it needs assisting mechanical forces to be activated, and other viral molecules may help it. They also found that nsp13 did not act like a Hepatitis C virus helicase with a similar shape, and instead acted more like ring-shaped helicases found in bacteriophages (viruses that infect bacteria).
Because coronaviruses have helicases that are very similar to nsp13, it is valuable to understand how this molecule works. “If we can come up with viral therapeutics that hit nsp13, we can have a first line of defense when new coronaviruses potentially erupt and cause new epidemics or pandemics in the future. Understanding this mechanism now can help us design inhibitors that can be treatments against coronaviruses,” Mickolajczyk says.
Their results, Mickolajczyk says, can provide insights that can be leveraged for drug discovery efforts. By inhibiting nsp13, a drug could prevent coronaviruses from making copies of themselves, thereby halting infections and stopping or preventing a pandemic.
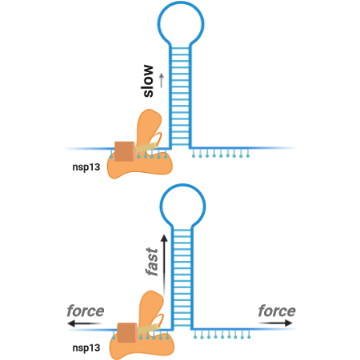
Nsp13 helicase (orange) unwinds RNA (blue), the same material as the SARS-CoV-2 genome. Recent findings show that mechanical forces (black arrows) increase nsp13 unwinding efficiency. Image courtesy of Keith Mickolajczyk.