SAN DIEGO, CA – Even though they don’t have conventional noses, insects have adapted to smell odors in nearly every imaginable niche. Mosquitoes find us by our odor molecules binding to odor receptors on their antennae, bees are drawn to flowers the same way, whereas ticks detect an approaching host using receptors on their forelegs. Insects’ ability to smell is uniquely adapted to their needs and habitats and Vanessa Ruta, Associate Professor at Rockefeller University, reveals a key to this versatility in research presented on Monday, February 17 at the 64th Annual Meeting of the Biophysical Society in San Diego, California.
“All animals use large families of olfactory receptors to detect the incredible variety of chemicals that exist around us. Insects evolved a unique molecular solution, the largest and most diverse group of specialized channels in nature, due to the large number of different insect species. And yet, we knew so little about these proteins,” Ruta said. Ruta and colleagues unveiled the structure of a protein for insect scent that provides an explanation for how insects evolved millions of odor receptors suited for a wide range of lifestyles and habitats.
Each insect olfactory unit is a channel comprised of two different proteins: an odor receptor that is opened by a specific scent molecule, and a co-receptor named Orco. While the odor receptors are highly specific to each species of insect, Orco is nearly the same in all of them. “You can take an Orco from one insect and it works with any other insect’s odor receptor,” explains Ruta. How one Orco assembles with hundreds of thousands of odor receptors was a fascinating evolutionary and molecular puzzle for Ruta, and she wanted to look at it in greater detail.
Using cryo-electron microscopy, Ruta and colleagues solved Orco’s structure, which serves as the first structural template to understand this huge protein family. “Orco itself can form an ion channel, unlike any others,” said Ruta. It features a central pore surrounded by four loosely-associated subunits, which explains how it can assemble with so many different odor receptors. “By minimizing inter-subunit interactions, it’s able to accommodate many protein sequences,” Ruta explained, they believe it can assemble with an odor receptor and share a common channel. Orco’s shape may also be crucial for evolution.
When Ruta mapped the sequence of odor receptors onto the Orco channel the similarity was strong in pore, but weak in the four loosely-associated subunits, which may allow those parts to diversify and evolve rapidly. Orco also likely provides a channel that works no matter how much the odor receptor diversifies, since they assemble together.
Most insects rely heavily on smells to navigate the world. And when it comes to insect vectors of disease, like mosquitoes, Ruta says, “this could be a starting point for designing insect repellents or insect confusants, that scramble the olfactory code and prevent them from finding their human hosts.”
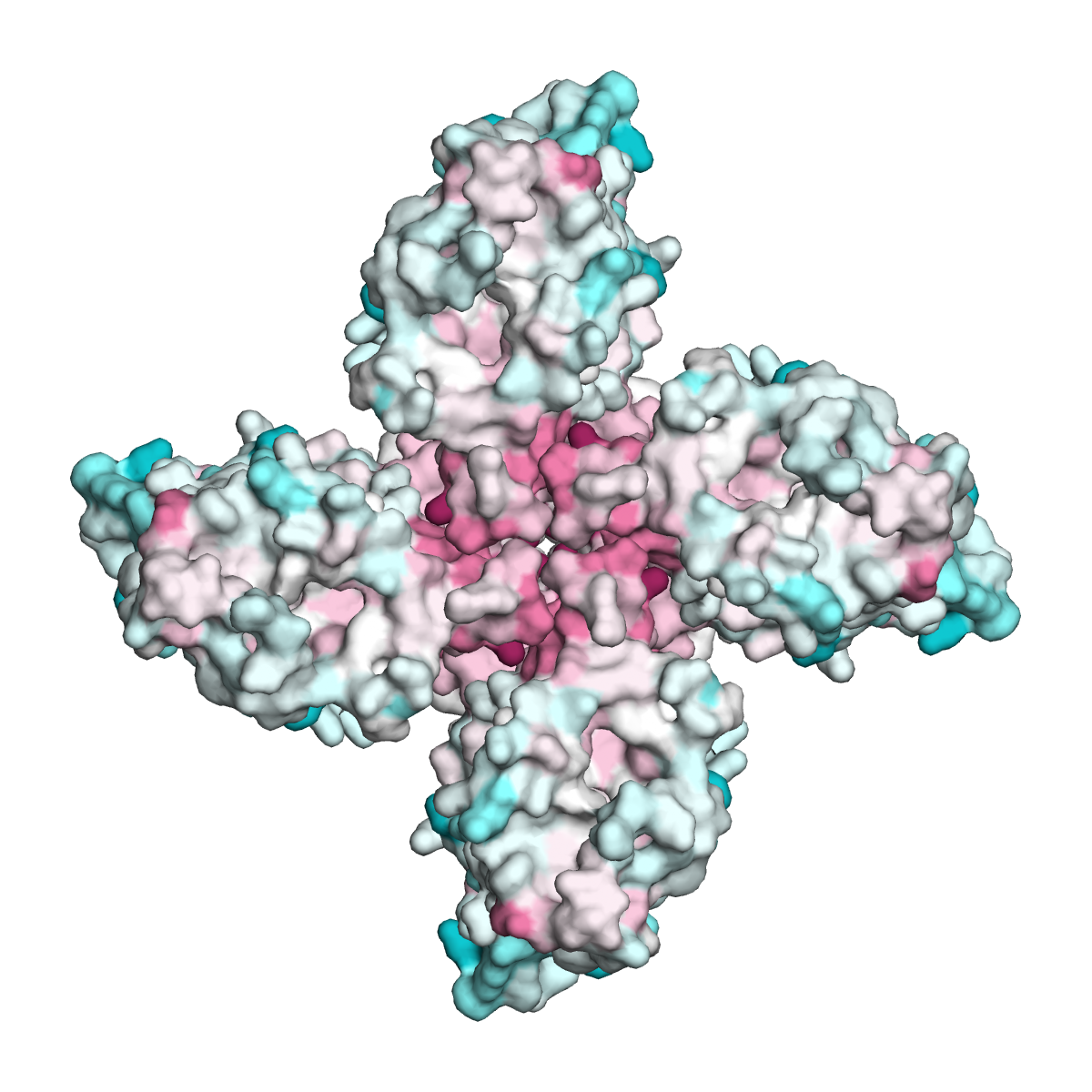
A view of the Orco channel from above, looking down into the pore. When odorant receptor sequences were mapped onto this Orco channel structure, some areas were highly similar, shown in magenta, and other areas were variable, shown in cyan. Most similarities in odorant receptors are concentrated along the central pore, leaving the rest of the protein free to diversify, suggesting how the architecture of this receptor family accommodates its remarkable sequence diversity and facilitates the evolution of odor tuning. Image courtesy of Vanessa Ruta.