Viruses and their infection routes have been hot topics for decades. Research into the mechanics of viral packaging and the roles of molecules involved in the process informs the development of effective diagnostic and treatment strategies. With the ongoing COVID-19 pandemic, the public knowledge of RNA viruses (viruses that have RNA as the genetic material) has risen. Another well-known RNA virus is HIV-1, which can severely damage the immune system and lead to acquired immune deficiency syndrome.
Over the years, scientists have shown that different viruses (and sometimes subtypes of the same virus) can have vastly different molecular mechanisms. In particular, the mechanism involved in viral RNA packaging can be linked to the infectiousness of the virus. Now, in a study reported in the November 2, 2021 issue of Biophysical Journal, scientists provide new information on how the sequences of a specific portion of the viral genome, the 5' untranslated region (UTR), play a role in the dimerization of HIV1 RNA genomes within viral capsids. Results from this article, “Stability and conformation of the dimeric HIV-1 genomic RNA 5ʹUTR,” could lead to a unified model of genome dimerization for different HIV-1 strains.
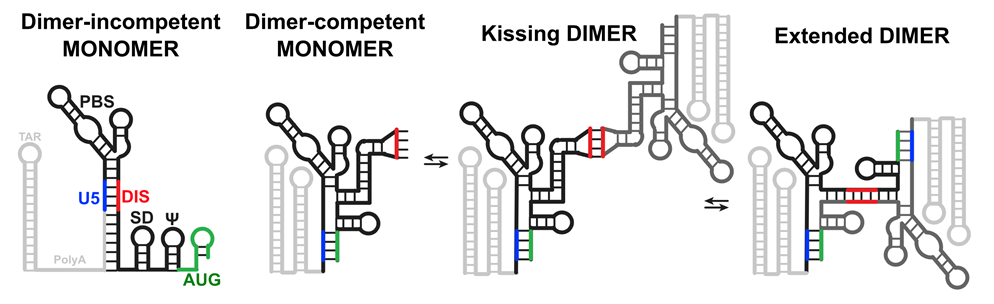
The HIV-1 viral “capsid” (the protein shell enclosing the genetic material) contains two copies of an RNA genome that are already dimerized when packaged into the capsid. Previous research had shown that this dimerization of the RNA genome can occur due to several factors such as the secondary structure of the 5'-UTR region and a specific six-nucleotide palindromic dimerization-initiation sequence (DIS) within the 5'-UTR. The new study by Blakemore and colleagues used two HIV-1 strains called MAL (sub-type A) and NL4.3 (sub-type B) to understand the roles of the structure and sequence of the 5'-UTR in HIV1 genome dimerization and whether certain features can be generalized for diverse subtypes of the virus. The team conducted studies on both subtypes and found that the 5'-UTR dimers from NL4.3 were stable at higher temperatures of up to 57°C, even in the absence of magnesium, whereas the MAL dimers were stable only when magnesium was present in the buffer. The team also identified the role of the DIS by swapping the DIS sequence of NL4.3 and MAL. They found that the stability of the dimers was a feature of the DIS sequence, with the MAL dimer now stable without magnesium because it contained the NL4.3 DIS sequence (and vice versa).
Through this study, the team of researchers shows that phenotypic differences in 5'-UTR dimer stability were based on sequence variation in the DIS stem loops. Their work could lead to a unified model for studying the dimerization of the genomes of diverse HIV-1 strains.